Lagging strand a closer look – Unraveling the intricacies of DNA replication, we delve into the fascinating world of the lagging strand. Embark on a journey to understand its unique mechanisms, challenges, and the intricate interplay of enzymes that ensure the faithful duplication of our genetic blueprint.
Lagging strand synthesis, a complex process in its own right, presents a myriad of challenges that require specialized enzymes and precise coordination. Join us as we explore the intricacies of this essential aspect of DNA replication, examining its impact on genome stability and the remarkable diversity observed across different organisms.
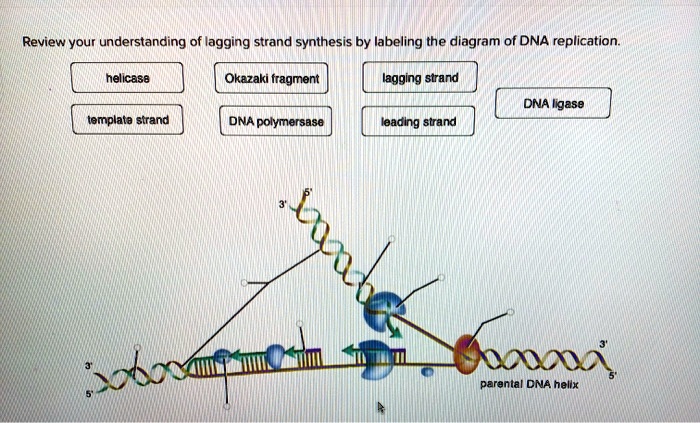
DNA Replication on the Lagging Strand

DNA replication is a fundamental process in cell biology that ensures the faithful transmission of genetic information from one generation to the next. It is a semi-conservative process, meaning that each newly synthesized DNA molecule consists of one original strand and one newly synthesized strand.On
the leading strand, DNA polymerase synthesizes DNA continuously in the 5′ to 3′ direction, following the unwound DNA template. However, on the lagging strand, DNA synthesis is more complex due to the antiparallel nature of the DNA double helix.
Okazaki Fragments
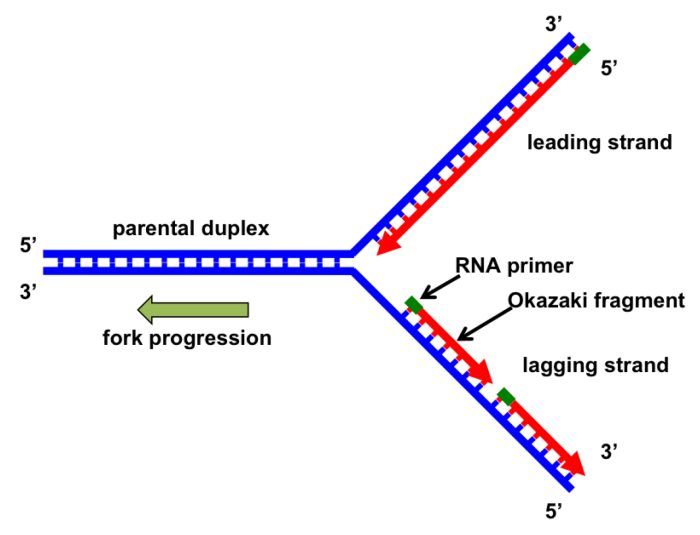
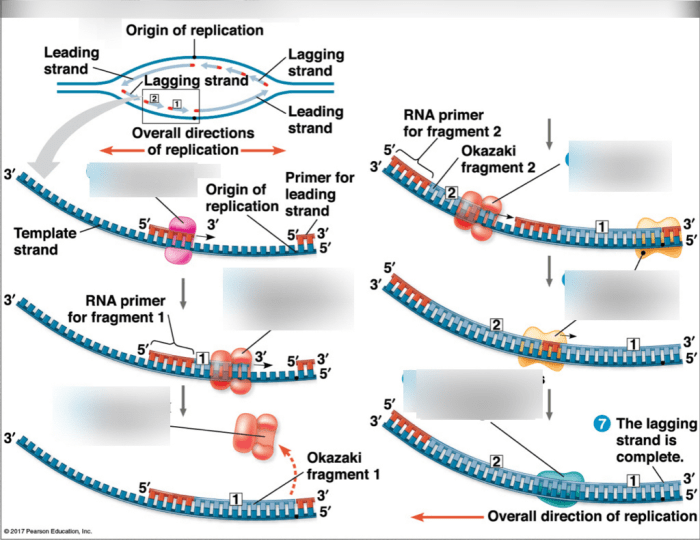
DNA polymerase can only synthesize DNA in the 5′ to 3′ direction. On the lagging strand, the DNA template is oriented in the opposite direction, so DNA polymerase must synthesize DNA in short fragments, called Okazaki fragments, in the 5′ to 3′ direction away from the replication fork.Once
an Okazaki fragment is synthesized, DNA ligase joins it to the previously synthesized fragment, creating a continuous strand of DNA. This process continues until the entire lagging strand is synthesized.
Comparison of Leading and Lagging Strand Synthesis
The following table compares the synthesis of the leading and lagging strands:| Feature | Leading Strand | Lagging Strand ||—|—|—|| Direction of synthesis | 5′ to 3′ | 5′ to 3′ in Okazaki fragments || Continuous or discontinuous | Continuous | Discontinuous || Role of DNA polymerase | Synthesizes DNA continuously | Synthesizes DNA in Okazaki fragments || Role of DNA ligase | Not required | Joins Okazaki fragments |
Enzymes Involved in Lagging Strand Synthesis
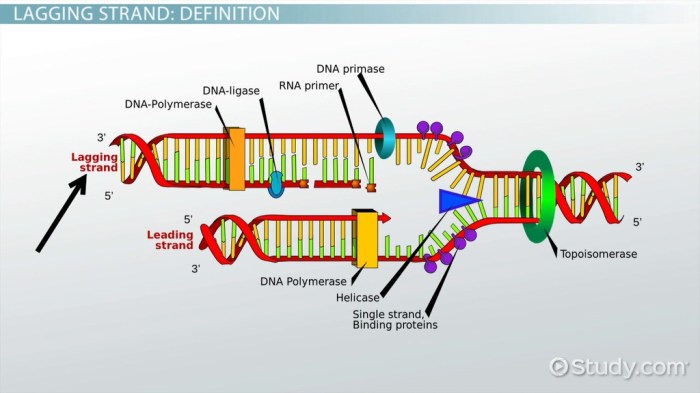
The lagging strand is synthesized in short fragments called Okazaki fragments. Several enzymes are involved in this process, including DNA polymerase III, DNA polymerase I, and ligase.
DNA polymerase III is the main enzyme responsible for synthesizing DNA on both the leading and lagging strands. It adds nucleotides to the 3′ end of the growing DNA strand, using the template strand as a guide.
DNA polymerase I is involved in several processes during DNA replication, including repairing errors made by DNA polymerase III and synthesizing the RNA primers that are required for DNA polymerase III to begin synthesis.
Ligase is an enzyme that joins the Okazaki fragments together to form a continuous DNA strand.
Lagging strand a closer look can sometimes feel like a chore, but it’s essential to understand the complexities of DNA replication. If you’re feeling overwhelmed, check out this article that compares it to feeling like a limp dishcloth. Lagging strand a closer look will make more sense when you can relate it to something you’ve experienced.
Coordination of Enzymes
These enzymes work together in a coordinated manner to ensure accurate replication of the DNA. DNA polymerase III is responsible for the majority of DNA synthesis, while DNA polymerase I and ligase play important roles in ensuring that the replication process is accurate and efficient.
Challenges and Errors in Lagging Strand Synthesis
Lagging strand synthesis faces challenges due to the discontinuous nature of the process and the need to displace the template strand during primer extension. Errors can also occur during this process, leading to mutations in the newly synthesized DNA.
Template Strand Displacement, Lagging strand a closer look
During lagging strand synthesis, the DNA polymerase must displace the template strand to extend the primer. This can be challenging, especially in regions with strong secondary structures or DNA-binding proteins. The polymerase can stall or even dissociate from the template, leading to gaps in the newly synthesized strand.
Errors in Lagging Strand Synthesis
Errors in lagging strand synthesis can occur due to misincorporation of nucleotides by the DNA polymerase, strand breaks, or incomplete primer extension. These errors can lead to mutations in the newly synthesized DNA, which can have various consequences, including altered protein function, cell cycle arrest, or even cell death.
Mechanisms to Prevent and Correct Errors
To prevent and correct errors in lagging strand synthesis, several mechanisms are employed by the cell. These include:
- Proofreading activity of the DNA polymerase
- Exonuclease activity of the DNA polymerase
- Mismatch repair system
These mechanisms work together to ensure the accuracy of DNA replication and minimize the risk of mutations.
Lagging Strand Synthesis in Different Organisms
Lagging strand synthesis exhibits variations among different organisms. Understanding these differences provides insights into the complexities and adaptations of DNA replication across diverse species.
Prokaryotes vs. Eukaryotes
Prokaryotes possess a circular chromosome, while eukaryotes have multiple linear chromosomes. This difference influences lagging strand synthesis. In prokaryotes, the circular chromosome allows for continuous synthesis of both leading and lagging strands. In eukaryotes, the linear chromosomes require the synthesis of multiple Okazaki fragments on the lagging strand, which are later joined by DNA ligase.
Genome Size and Replication Time
The number of Okazaki fragments generated during lagging strand synthesis affects genome size and replication time. Organisms with smaller genomes, such as bacteria, have fewer Okazaki fragments and a faster replication time compared to organisms with larger genomes, such as eukaryotes.
The larger number of Okazaki fragments in eukaryotes requires more time for their synthesis and ligation, contributing to the longer replication time.
Unique Lagging Strand Synthesis Mechanisms
Some organisms have evolved unique mechanisms for lagging strand synthesis. For example, in the archaeon Sulfolobus solfataricus, the lagging strand is synthesized by a specialized DNA polymerase, Dpo4, which can bypass certain DNA lesions. In the bacterium Thermus aquaticus, the lagging strand is synthesized by a discontinuous mechanism involving multiple DNA polymerases and a unique protein complex.
Popular Questions: Lagging Strand A Closer Look
What is the key difference between leading and lagging strand synthesis?
Leading strand synthesis proceeds continuously in the 5′ to 3′ direction, while lagging strand synthesis occurs discontinuously in the form of Okazaki fragments.
What is the role of DNA polymerase III in lagging strand synthesis?
DNA polymerase III is the primary enzyme responsible for synthesizing new DNA strands on both the leading and lagging strands.
How are errors prevented and corrected during lagging strand synthesis?
DNA polymerase III has proofreading capabilities to correct errors during synthesis, while DNA polymerase I removes RNA primers and fills in the gaps between Okazaki fragments.